| structure name | CRYSTAL STRUCTURE OF MAD-MAX RECOGNIZING DNA |
| reference | Burley et al. 'Cell(Cambridge,Mass.)' 112 193 2003 |
| source | HOMO SAPIENS |
| experiment | X-ray (resolution=2.00, R-factor=0.264) |
| structural superfamily | HLH, helix-loop-helix DNA-binding domain; |
| sequence family | Helixloophelix DNAbinding domain; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | HNERR |
Estimated binding specificities ?
| contact | 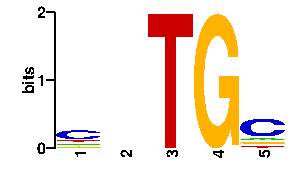 |
A | 12 0 96 24 14 C | 12 96 0 24 14 G | 60 0 0 24 54 T | 12 0 0 24 14scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: MAD | ||
| CACGTG | PubMed | 1nlw_AB(3.65e-09) |
| CATGTG | PubMed | 1nlw_AB(1.66e-04) |
Dendrogram of similar interfaces ?
matrix format----HGNT----EC----RCRG---- L +--1nkp_A +-----4 ----HGNT----EC----RCRC---- L ! ! +1nlw_A ! +-1 ----HGNT----EC----RCRG---- L ! +1nkp_E +---------------7 --RC----HTNC--IGEC----YGRC L ! ! +1am9_D ! ! +------3 ----HGNT----EC----RCRG---- L ! ! ! +1hlo_A ! +-6 ------NT----ET----RCMT---- --8 ! +--2ql2_D ! +----5 ------NT--RGEC----RC------ L ! ! +2ql2_A ! +-2 RG----TT--RAEC----RC------ ! +1mdy_B ! RC--HG------EA----RARG---- L +------------------------1a0a_A |
home
updated Mon Dec 18 05:31:26 2023
