| structure name | PROTELOMERASE TELK COMPLEXED WITH SUBSTRATE DNA (PROTELEMORASE) |
| reference | Ellenberger et al. Mol.Cell 27 901 2007 |
| source | KLEBSIELLA PHAGE PHIKO2 |
| experiment | X-ray (resolution=3.20, R-factor=0.255) |
| multimeric complexes | 2v6e_AB |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | NTNSYSRRQWRKRRNAATNDTHSRTR |
Estimated binding specificities ?
| readout + contact | 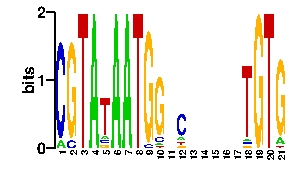 |
A | 3 96 0 84 24 24 24 24 24 11 24 7 1 96 0 0 77 0 96 0 0 C | 77 0 96 3 24 24 24 24 24 11 24 70 92 0 0 0 3 0 0 88 0 G | 3 0 0 6 24 24 24 24 24 63 24 11 3 0 0 0 7 0 0 7 89 T | 13 0 0 3 24 24 24 24 24 11 24 8 0 0 96 96 9 96 0 1 7scan! |
Dendrogram of similar interfaces ?
matrix formatNTTTNTSAYTSTRT----RG--QCWT----RG--KA--RGRC----------NAAC----AATG----NT--DG--TG--HA--SG--RGTCRT-------- L +------------------2v6e_B +-----8 ------------------YA--RAKT--------KA--------IT------ST------RTLT----RT--DAEATT------------------------ L ! ! +4f43_A ! +------------------1 --------------TAAAYADA--KA----------------IT------ST------RTLT----RT---------------------------------- L ! +4e0p_A +-11 ----------------------------------------------------------YG--YAKT------------TT---------------------- L ! ! +-----------5jk0_B ! ! +----------5 ----------------------------------------------ATVTDT------------RC------------------------------------ L +-12 +-10 +-----------6en2_A ! ! ! ------------------------------------KT--------------------TT--STRGYC---------------------------------- L ! ! +----------------------1z1b_B +-13 ! --------------------------RAFC--------------------------NT----RGST----KT------------------------------ L ! ! +--------------------------1flo_A ! ! --------------------------------NC----------------------------TT-------------------------------------- L ! +---------------------------2b9s_A ! ------------------------------------------------------------------------------------------------YG---- ! +-1sfu_A -14 +------------4 ------------------------------------------------------------------------------------------------YG---- ! ! +-4lb5_B ! +-6 ------------------------------------------------------------------------------------------------YG---- ! ! ! +2heo_D ! +-----7 +--------------2 ----------------------------------------------------------------------------------------------RCYG--KT ! ! ! +2acj_B ! ! ! ----------------------------------------------------------------------------------SG--QG--YCNC-------- +------9 +---------------2ve9_C ! ----------------------------------------------------------------------------------------------RGYG---- ! +-1qbj_C +-------------------3 ------------------------------------------------------------------------------------------------YGRG-- +-4wcg_A |
home
updated Tue Dec 19 06:58:01 2023
