| structure name | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH DNA AND CA (HOMING ENDONUCLEASE I-DMOI) |
| reference | Marcaida et al. Proc.Natl.Acad.Sci.USA 105 16888 2008 |
| source | DESULFUROCOCCUS MOBILIS |
| experiment | X-ray (resolution=2.05, R-factor=0.189) |
| structural superfamily | Homing endonucleases; |
| redundant complexes | 2vs8_K
 4d6n_F
4d6o_A
4una_A
4unb_G
4unc_G
4ut0_A
5a0w_A
5ak9_A
5akf_I
5akm_A
5akn_K
5o6g_A
5o6i_K 4d6n_F
4d6o_A
4una_A
4unb_G
4unc_G
4ut0_A
5a0w_A
5ak9_A
5akf_I
5akm_A
5akn_K
5o6g_A
5o6i_K
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | YLYRSERVTQVTRERSRRWDDRHV |
Estimated binding specificities ?
| readout + contact | 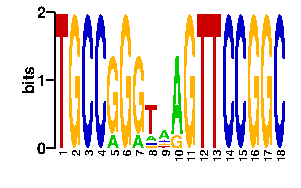 |
A | 0 0 0 0 0 96 96 0 1 9 67 0 0 0 0 0 0 96 C | 0 96 96 0 0 0 0 96 9 19 9 87 96 86 0 0 96 0 G | 96 0 0 96 96 0 0 0 0 9 9 0 0 0 96 96 0 0 T | 0 0 0 0 0 0 0 0 86 59 11 9 0 10 0 0 0 0scan! |
Dendrogram of similar interfaces ?
matrix format--YTLT--YC----RGSGECRGVTTG----QG--VG------TARTEARGSC--RGRGWCDCDCRGHAVC L +---------2vs7_A +-------------7 YGRGRG----------------QGEC--------------------------NT----------NCKT-- L ! +---------3c0w_A ! SANC--------------DAECNATG--RC--KGST--------------DA------------------ L ! +-------2ex5_A ! ! STIT----------QT--ST--ATTC--------YT--SC----VCDTRGST------------------ L ! +------6 +3mxb_B -10 ! ! +-1 SGIG----------RG--QG--DGTA----YC--AT--SC----VCKGRG-------------------- L ! ! +-----4 +6fb0_A ! ! ! SAYCLC--------------------NA--RG----RG------IC--RG-------------------- L ! +----8 +-2fld_B ! ! ! CGRG--ST--TTKARG--QTFAQGNG------HC--SC--------QGRGQA------------------ L ! ! ! +4yit_A ! ! ! +-2 ST------------AT--LCYCECAT----YG--NA--SADC----KTKGSA------------------ L +---9 +-------------3 +4lq0_A ! ! STIGRG--------LT--EC--STKG------TT--NA--------MGRTSC------------------ L ! +5t2n_A ! CA----------------RT----FA----RG----RG--DT--MCEA---------------------- L ! +-5a74_B +-----------------5 SAIA----------RG--KG--QATA-------------------------------------------- L +-2vbl_B |
home
updated Tue Dec 19 06:59:48 2023
