| structure name | CRYSTAL STRUCTURE OF A UVRB DIMER-DNA COMPLEX (UvrABC system protein B) |
| reference | Webster et al. 'Nucleic 40 8743 2012 |
| source | Bacillus subtilis |
| experiment | X-ray (resolution=3.25, R-factor=0.182) |
| structural superfamily | P-loop containing nucleoside triphosphate hydrolases; |
| sequence family | Helicase conserved Cterminal domain; Type III restriction enzyme, res subunit; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | YPYQMRNFL |
Estimated binding specificities ?
| contact | 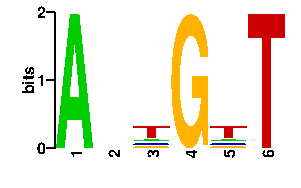 |
A | 96 24 16 0 13 0 C | 0 24 13 0 13 0 G | 0 24 13 96 16 0 T | 0 24 54 0 54 96scan! |
Dendrogram of similar interfaces ?
matrix formatYTPTYTQTMGRT--------NA--FALA-- L +-3v4r_B +-------------------5 ----------RG------------AG---- +-12 +-6o8h_A ! ! ------------RT---------------- L ! +---------------------6fws_A ! ----------------------------WA ! +6pwf_K ! +-2 ----------------HCKTRC-------- ! ! +7egp_A ! +-4 ----------------QT------------ -13 ! ! +6ne3_W ! +------------6 +-3 ----------------ST------------ ! ! ! +6jyl_K ! ! ! ----------------SGRT---------- ! +---10 +--7enn_K ! ! ! --------------------RT----DAWA L ! ! ! +--------5vvr_M ! ! +------8 --------------------RC-------- ! ! ! +----7nkx_W +--11 +---7 --------------------RC-------- ! +----6ryr_W ! --------------YT------YA------ ! +--------6p4f_C +-----------9 ------HG------------KG--HC---- ! +7nvv_7 +--------1 --------------------KG-------- +7o4j_7 |
home
updated Fri Sep 13 17:49:37 2024
