| structure name | STRUCTURAL BASIS FOR ARYL HYDROCARBON RECEPTOR MEDIATED GENE ACTIVATION (ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSLOCATOR) |
| reference | Schulte et al. Structure 25 1025 2017 |
| source | MUS MUSCULUS |
| experiment | X-ray (resolution=3.30, R-factor=0.294) |
| structural superfamily | HLH, helix-loop-helix DNA-binding domain; |
| sequence family | Helixloophelix DNAbinding domain; |
| multimeric complexes | 5nj8_AB 5v0l_AB 7xhv_AB |
| redundant complexes | 5v0l 7xhv |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | HERR |
Estimated binding specificities ?
readout + contact + contact | 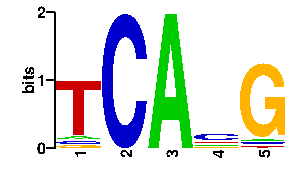 |
A | 4 15 0 0 78 C | 84 17 0 0 6 G | 4 49 0 96 6 T | 4 15 96 0 6scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: AHR | ||
| GCGTG | PubMed | 5nj8_D(1.35e-04), 5nj8_AB(1.62e-06) |
| CACGC | PubMed | 5nj8_D(1.35e-04), 5nj8_AB(1.62e-06) |
| GTGCGTG | PubMed | 5nj8_D(1.01e-03), 5nj8_AB(1.65e-07) |
Dendrogram of similar interfaces ?
matrix format------HG----ECRGRG L +-------------4zpk_A +------------3 ------HG----ETRTRG L ! +-------------5v0l_A ! ------HGSTITETRCRG L --5 +4h10_A ! +--1 KT----HGNT--ECRCRG L ! +-----------------2 +1nkp_A ! ! ! --RT----NTRGECRCQT +-----4 +--2ypa_A ! NTNGRTECRGRC------ L +--------------------6od3_F |
home
updated Sat Jan 10 08:29:39 2026
