| structure name | CRYSTAL STRUCTURE OF MUTM FROM NEISSERIA MENINGITIDIS (Formamidopyrimidine-DNA glycosylase) |
| reference | Landova et al. 'Febs 594 3032 2020 |
| source | Neisseria meningitidis alpha522 |
| experiment | X-ray (resolution=2.17, R-factor=0.240) |
| structural superfamily | N-terminal domain of MutM-like DNA repair proteins;S13-like H2TH domain;Glucocorticoid receptor-like DNA-binding domain; |
| sequence family | FormamidopyrimidineDNA glycosylase H2; FormamidopyrimidineDNA glycosylase N; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RMRFR |
Estimated binding specificities ?
| contact | 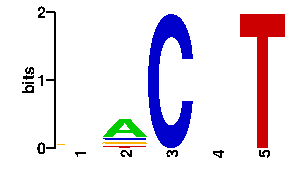 |
A | 19 60 0 24 0 C | 19 12 96 24 0 G | 38 12 0 24 0 T | 20 12 0 24 96scan! |
Dendrogram of similar interfaces ?
matrix formatRT--MG----RCFC--------RT L +--6tc9_A +-1 RA--MG----RCFCKGDGACQTRG L +--------2 +--1k82_A ! ! --RA------RGFT--------RA L +-----------3 +--3gpu_A ! ! --QALGYA--------------RG --4 +-----------1k3x_A ! ----LG--MTRCFC--------RT L +-----------------------3vk8_B |
home
updated Sat Jan 10 12:01:41 2026
