| structure name | CRYSTAL STRUCTURE OF THE INTEIN HOMING ENDONUCLEASE PI-SCEI BOUND TO ITS SUBSTRATE DNA (CA2+ FREE) (ENDONUCLEASE PI-SCEI) |
| reference | Quiocho et al. |
| source | Saccharomyces cerevisiae |
| experiment | X-ray (resolution=3.20, R-factor=?) |
| structural superfamily | Hedgehog/intein Hint domain;Homing endonucleases; |
| sequence family | Hom_endassociated Hint; Homing endonuclease; |
| redundant complexes |  1lws_A 1lws_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QRLRIKYEESSSHKRYKNEKDNTHY |
Estimated binding specificities ?
| readout + contact | 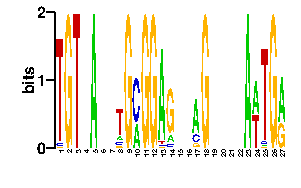 |
A | 1 0 0 24 96 24 24 9 0 38 0 0 94 9 24 24 58 0 24 24 24 24 96 38 1 0 67 C | 1 0 0 24 0 24 24 15 0 57 0 0 0 9 24 24 26 0 24 24 24 24 0 0 0 0 0 G | 4 96 0 24 0 24 24 11 96 1 96 96 0 73 24 24 9 96 24 24 24 24 0 1 3 96 29 T | 90 0 96 24 0 24 24 61 0 0 0 0 2 5 24 24 3 0 24 24 24 24 0 57 92 0 0scan! |
Dendrogram of similar interfaces ?
matrix formatQGRALARGITKAYTETECSTSTSTHTKGRG------YA----KG------NTECKCDGNCTTHG--YT L +---------------------1lwt_A +------2 ------------------------------QGEC--------QC------------------------ L ! ! +-------------3c0w_A --3 +-------1 --------------------------------TGQG--RGRGWC--------------------VC-- L ! +-------------2vs7_A ! --------------------------------------------KGCGFT------------------ L +----------------------------2bop_A |
home
updated Sun Aug 3 05:46:11 2025
