| structure name | CRYSTAL STRUCTURE OF AN IDER-DNA COMPLEX REVEALS A CONFORMATIONAL CHANGE IN ACTIVATED IDER FOR BASE-SPECIFIC INTERACTIONS |
| reference | Wisedchaisri et al. J.Mol.Biol. 342 1155 2004 |
| source | Mycobacterium tuberculosis |
| experiment | X-ray (resolution=2.75, R-factor=?) |
| structural superfamily | Iron-dependent repressor protein, dimerization domain;"Winged helix" DNA-binding domain; |
| reference complex | 2isz_B |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | SPTQR/SPTSQR/SPTQR/SPTQR |
Estimated binding specificities ?
| contact | 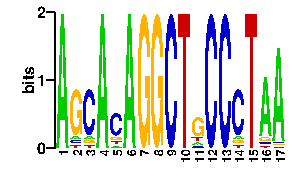 |
A | 2 9 96 11 0 0 7 96 0 0 0 0 12 0 8 5 0 C | 2 7 0 5 0 0 70 0 0 96 96 0 12 0 5 76 0 G | 2 10 0 75 96 96 12 0 96 0 0 0 60 0 78 5 0 T | 90 70 0 5 0 0 7 0 0 0 0 96 12 96 5 10 96scan! |
home
updated Sun Jan 4 23:48:50 2026
