| structure name | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE (GAGA-FACTOR) |
| source | DROSOPHILA MELANOGASTER |
| experiment | NMR |
| structural superfamily | beta-beta-alpha zinc fingers; |
| sequence family | GAGA factor; |
| redundant complexes | 1yuj |
| reference complex | 1yuj_A |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RKRSQRNR |
Estimated binding specificities ?
| readout + contact | 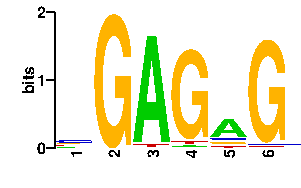 |
A | 16 0 90 3 60 1 C | 44 0 0 1 12 2 G | 18 96 2 88 12 91 T | 18 0 4 4 12 2scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: GAGA | ||
| GAGAG | PubMed | 1yui_A(1.20e-07), 1yuj_A(1.29e-07) |
| GAGAGGGAGGAG | PubMed | 1yui_A(1.61e-04), 1yuj_A(1.69e-04) |
| GAGAGAG | PubMed | 1yui_A(6.29e-06), 1yuj_A(6.68e-06) |
| GAGNGAG | PubMed | 1yui_A(3.37e-03), 1yuj_A(3.52e-03) |
| CCGAGAGG | PubMed | 1yui_A(1.76e-05), 1yuj_A(1.87e-05) |
| TGTCTC | PubMed | 1yui_A(1.12e-03), 1yuj_A(1.17e-03) |
| CGAGAC | PubMed | 1yui_A(1.12e-03), 1yuj_A(1.17e-03) |
| CCCTC | PubMed | 1yui_A(2.62e-03), 1yuj_A(2.74e-03) |
home
updated Thu Jan 8 19:30:29 2026
