| structure name | CRYSTAL STRUCTURE OF XERH SITE-SPECIFIC RECOMBINASE BOUND TO DIFH SUBSTRATE: PRE-CLEAVAGE COMPLEX (Tyrosine recombinase XerH) |
| reference | Barabas et al. Elife 5 ? 2016 |
| source | Helicobacter pylori (strain ATCC 700392 / 26695) / Helicobacter pylori (strain ATCC 700392 / 26695) / Helicobacter pylori (strain ATCC 700392 / 26695) |
| experiment | X-ray (resolution=2.10, R-factor=0.193) |
| structural superfamily | DNA breaking-rejoining enzymes; |
| sequence family | Phage integrase family; |
| redundant complexes |  5jjv_B 5jjv_B
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RNLNTYATKNITQAYYKT |
Estimated binding specificities ?
| readout + contact | 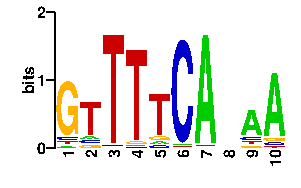 |
A | 5 9 1 2 8 2 92 23 65 80 C | 7 10 2 2 7 91 1 23 11 6 G | 78 8 1 4 10 1 1 16 9 6 T | 6 69 92 88 71 2 2 34 11 4scan! |
Dendrogram of similar interfaces ?
matrix formatRGNTLTNTTTYT--AATAKGNAIT------TTQAATYGYAKT--------TT L +------------5jk0_B +---------2 --------------KTTT--NARASAYT----ATVTDT--------NT--MT L ! +------------6en1_A +-4 ------------TA--------------------RTLT--RT--NADAEATT L ! ! +4f43_A +----5 +----------------------1 ----------------------------------RT----RT----DAEATT L ! ! +4e0g_A ! ! --------------ATST--LT------KT------TTSTRGYC-------- L --6 +-----------------------1z1b_B ! ------------------------------RT--ET---------------- ! +-------------6gys_E +--------------3 ----------------------------------LT---------------- +-------------3t79_A |
home
updated Fri Sep 13 19:56:38 2024
