| structure name | CRYO-EM STRUCTURE OF THE MVV CSC INTASOME AT 4.5A RESOLUTION |
| source | VISNA/MAEDI VIRUS EV1 KV1772 |
| experiment | NMR |
| structural superfamily | Ribonuclease H-like;N-terminal Zn binding domain of HIV integrase;DNA-binding domain of retroviral integrase; |
| reference complex | 7u32_A |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RESR/AKA/QARQ/TNKP |
Estimated binding specificities ?
| contact | 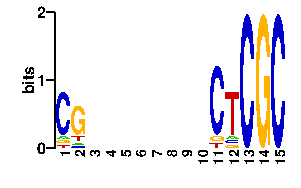 |
A | 0 0 0 73 7 24 24 24 24 24 24 24 24 7 7 C | 0 96 0 7 9 24 24 24 24 24 24 24 24 75 7 G | 96 0 96 7 73 24 24 24 24 24 24 24 24 7 75 T | 0 0 0 9 7 24 24 24 24 24 24 24 24 7 7scan! |
home
updated Wed Jan 7 18:05:42 2026
