| structure name | MVV CLEAVED SYNAPTIC COMPLEX (CSC) INTASOME AT 3.4 A RESOLUTION (INTEGRASE) |
| source | VISNA/MAEDI VIRUS EV1 KV1772 |
| experiment | NMR |
| structural superfamily | Ribonuclease H-like;N-terminal Zn binding domain of HIV integrase;DNA-binding domain of retroviral integrase; |
| sequence family | Integrase core domain; Integrase DNA binding domain; Integrase Zinc binding domain; |
| multimeric complexes | 5m0r_AI 5m0r_DN 5m0r_FL 7u32_AFL 7u32_DIN 7z1z_DEIN 7z1z_FL 7zpp_ADIN 7zpp_FL |
| redundant complexes | 5m0r_N 7z1z_M 7zpp_A |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QAERQ |
Estimated binding specificities ?
| contact | 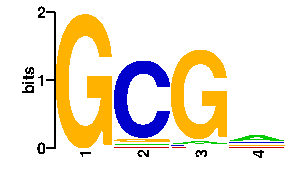 |
A | 19 2 5 0 C | 16 88 3 96 G | 16 4 85 0 T | 45 2 3 0scan! |
Dendrogram of similar interfaces ?
matrix formatHG--DT----------------QGECSGHCQT---------- +-----------------------------6rwm_A +-2 ------KT--------HTETPAQAEARC-------------- L ! ! +----------------3oym_A +-3 +------------1 HGQTDT--TAHA----------QGECSCKG------------ ! ! +----------------6vdk_A --4 ! --------------------------------RAYACAPAWA ! +-----------------------------3jca_H ! ------------KTYC-------------------------- L +-----------------------------6x68_D |
home
updated Sat Jan 10 17:34:51 2026
