| structure name | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA- DEPENDENT RNA POLYMERASE (RNA-DIRECTED RNA POLYMERASE) |
| reference | Koivunen et al. 'Nucleic 36 6633 2008 |
| source | PSEUDOMONAS PHAGE PHI6 |
| experiment | X-ray (resolution=2.80, R-factor=0.220) |
| structural superfamily | DNA/RNA polymerases; |
| sequence family | RNA dependent RNA polymerase; |
| redundant complexes | 1hht_R
 4a8s_C 4a8s_C
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QRKVRAQRSTYKLQTEH |
Estimated binding specificities ?
| contact | 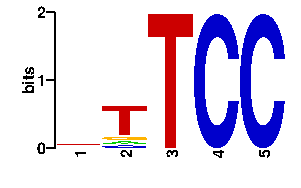 |
A | 19 9 0 0 0 C | 19 9 0 96 96 G | 19 11 0 0 0 T | 39 67 96 0 0scan! |
Dendrogram of similar interfaces ?
matrix formatQTRTKCVTRT----AT--QCRC--STTC--------YTKTLCQCTCECHC L +--------------------2jlg_B +-----1 ----------------------------YA-------------------- +-2 +--------------------6ar1_A ! ! ------------------------------KGLGKG-------------- L --3 +--------------------------6me0_C ! ----------LCIC--FA----YG-------------------------- +---------------------------7kqn_D |
home
updated Tue Dec 17 22:53:10 2024
