| structure name | ATOMIC STRUCTURE OF THE POXVIRUS TRANSCRIPTION PRE-INITIATION COMPLEX IN THE INITIALLY MELTED STATE |
| source | VACCINIA VIRUS GLV-1H68 |
| experiment | NMR |
| structural superfamily | P-loop containing nucleoside triphosphate hydrolases; |
| reference complex | 7amv_W |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | K/STATNISIKYSRQKY/KFKSK |
Estimated binding specificities ?
| contact | 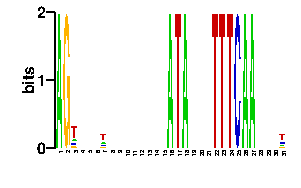 |
A | 38 31 27 24 12 1 9 70 78 52 24 24 24 4 96 0 24 24 24 24 24 24 24 24 31 24 22 24 34 0 0 C | 19 21 25 24 12 1 11 8 6 14 24 24 24 2 0 0 24 24 24 24 24 24 24 24 23 24 30 24 20 96 0 G | 19 23 22 24 12 1 67 8 6 16 24 24 24 2 0 0 24 24 24 24 24 24 24 24 21 24 22 24 22 0 0 T | 20 21 22 24 60 93 9 10 6 14 24 24 24 88 0 96 24 24 24 24 24 24 24 24 21 24 22 24 20 0 96scan! |
home
updated Tue Jan 6 20:09:59 2026
