| structure name | THE PHAGE 434 OR2/R1-69 COMPLEX AT 2.5 ANGSTROMS RESOLUTION |
| reference | Shimon et al. J.Mol.Biol. 232 826 1993 |
| experiment | X-ray (resolution=2.50, R-factor=0.209) |
| structural superfamily | lambda repressor-like DNA-binding domains; |
| sequence family | Helixturnhelix; |
| multimeric complexes | 1per_LR 1rpe_LR 2or1_LR |
| redundant complexes | 1per 2or1 |
| reference complex | 1per_L |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | TQQSEQ |
Estimated binding specificities ?
| contact | 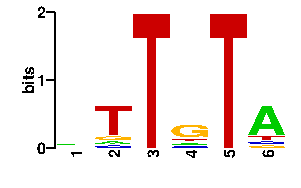 |
A | 38 9 0 13 0 67 C | 19 9 0 13 0 9 G | 20 11 0 56 0 9 T | 19 67 96 14 96 11scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: 434 REPRESSOR | ||
| CATACAAGAAAGNNNNNNTTT | PubMed | 1rpe_R(8.40e-04), 2or1_R(8.44e-04), 1per_L(8.44e-05) |
| ACAAGA | PubMed | 1rpe_R(1.12e-03), 2or1_LR(3.76e-04), 2or1_R(1.12e-03), ... (total=4) |
| ACAGTTTTCTTGT | PubMed | 2or1_LR(1.49e-03), 1per_L(1.02e-02) |
| ACAAAACTTTCTTGT | PubMed | 2or1_LR(1.49e-04) |
Dendrogram of similar interfaces ?
matrix format--QT--STNTVTATSTQTRTETTT-- L +--2r1j_L +-1 RT--------RG----NTET--RTYA L +------------------2 +--7f9h_B ! ! ----AT--QAAT--SA---------- +------3 +---4z5h_A ! ! --------QASTCTSART--ET---- L --4 +----------------------5j2y_A ! --QT----VTNT--STRG-------- L +-----------------------------3o9x_A |
home
updated Thu Jan 8 19:03:32 2026
