| structure name | 1H, 13C AND 15N CHEMICAL SHIFT ASSIGNMENTS AND SOLUTION STRUCTURE FOR PARP-1 F1F2 DOMAINS IN COMPLEX WITH A DNA SINGLE-STRAND BREAK (POLY ADP-RIBOSE POLYMERASE 1) |
| source | HOMO SAPIENS |
| experiment | NMR |
| structural superfamily | Glucocorticoid receptor-like DNA-binding domain; |
| sequence family | PolyADPribose polymerase and DNALigase Z; |
| multimeric complexes | 4av1_ABCD 4dqy_AD 4dqy_BF 4dqy_CE 4opx_AD 4oqa_AD 4oqb_AD 7s6h_AB 7s6m_CD 8g0h_CD |
| redundant complexes | 3odc_B 3ode_B 4av1_B 4opx_F 4oqa_F 4oqb_C 7s6h_D 8g0h_B 4dqy 7s6m |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RSFVPRQLI |
Estimated binding specificities ?
| contact | 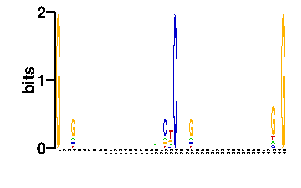 |
A | 0 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 38 24 12 14 0 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 21 9 24 0 C | 0 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 19 24 60 14 96 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 33 9 24 0 G | 96 24 24 60 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 19 24 12 16 0 24 24 60 24 24 24 24 24 24 24 24 24 24 24 24 24 24 21 67 24 96 T | 0 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 20 24 12 52 0 24 24 12 24 24 24 24 24 24 24 24 24 24 24 24 24 24 21 11 24 0scan! |
Dendrogram of similar interfaces ?
matrix format--RGSTFGVTPCRG--QCLCIG L +------------2n8a_A +--------1 ------------RGKG--LCIG +--2 +------------3odc_B ! ! --RG--FCVGPG---------- --3 +---------------------3od8_G ! STRG------------------ +------------------------4av1_A |
home
updated Sat Jan 10 05:24:57 2026
