| structure name | STRUCTURE OF THE AVRBS3-DNA COMPLEX PROVIDES NEW INSIGHTS INTO THE INITIAL THYMINE-RECOGNITION MECHANISM (AVRBS3) |
| reference | Yefimenko et al. 'Acta 69 1707 2013 |
| source | XANTHOMONAS CAMPESTRIS |
| experiment | X-ray (resolution=2.55, R-factor=0.239) |
| structural superfamily | Thiolase-like; |
| sequence family | Xanthomonas avirulence protein, Avr/PthA; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RDIIIIDDIIDDDD |
Estimated binding specificities ?
readout + contact + contact | 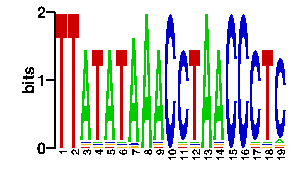 |
A | 0 0 90 2 88 2 88 96 88 0 2 2 96 88 0 0 2 2 5 C | 0 0 2 2 2 4 4 0 4 96 90 2 0 4 96 96 90 4 85 G | 0 0 2 4 4 2 2 0 2 0 2 2 0 2 0 0 2 2 2 T | 96 96 2 88 2 88 2 0 2 0 2 90 0 2 0 0 2 88 4scan! |
Dendrogram of similar interfaces ?
matrix formatRT--RTDA----IA--ITIAIADCDC--ITIADCDCDC--DC L +--2ypf_A +---1 --IAIANGTG--NGIAIANGDCIAIATARGSADCNG----SA L +--------2 +--4cja_A ! ! ----RADCIA----DC----DCDCIC----IADC----IAIC L --3 +------3ugm_A ! ----RADCDCHTDC------IA--DC--DC------------ L +---------------4osh_B |
home
updated Sat Jan 10 05:43:12 2026
