| structure name | CORE MODEL OF THE MOUSE MAMMARY TUMOR VIRUS INTASOME (INTEGRASE) |
| source | MOUSE MAMMARY TUMOR VIRUS |
| experiment | NMR |
| structural superfamily | Ribonuclease H-like;DNA-binding domain of retroviral integrase;N-terminal Zn binding domain of HIV integrase; |
| sequence family | Integrase core domain; Integrase DNA binding domain; Integrase Zinc binding domain; |
| multimeric complexes | 7ut1_ABG 7ut1_CE |
| redundant complexes | 7usf_D 7ut1_d |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QARQR |
Estimated binding specificities ?
| contact | 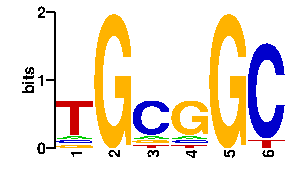 |
A | 8 0 11 8 0 0 C | 8 0 69 11 0 87 G | 11 96 8 69 96 1 T | 69 0 8 8 0 8scan! |
Dendrogram of similar interfaces ?
matrix formatVT--PT----------PAQAAGEARCQG--------RG-------- L +--3jca_E +----------------1 --NT------WA--TA--QGAGECRTRC--ET----RGYT------ L +---3 +--7jn3_E ! ! HG----DT----------QG--ECSGHCQT---------------- +--4 +-------------------6rwm_A ! ! HG--QT--DT--------QG--ECSCKG------------------ ! ! +---6vdk_A +-5 +-------------------2 ------------QT--PAQGAGECRGQG------------------ L ! ! +---7u32_A --6 ! ------------------------------------RAYACAPAWA ! +--------------------------3jca_H ! --------------------------------LAWAETACVAIA-- +---------------------------6puy_D |
home
updated Sat Jan 10 06:09:40 2026
