| structure name | CRYO-EM STRUCTURE OF HTLV-1 INSTASOME |
| experiment | NMR |
| structural superfamily | Ribonuclease H-like;DNA-binding domain of retroviral integrase; |
| sequence family | Integrase core domain; Integrase DNA binding domain; Integrase Zinc binding domain; |
| multimeric complexes | 6voy_AD 6voy_BC |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | IRPTQRRLKW |
Estimated binding specificities ?
| contact | 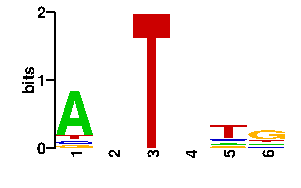 |
A | 74 24 0 21 13 14 C | 7 24 0 21 14 14 G | 7 24 0 33 13 52 T | 8 24 96 21 56 16scan! |
Dendrogram of similar interfaces ?
matrix format----IT--RT----------PATA--QCRG--------RALAKA----------WA L +6voy_C +-----------------1 --------RT----------PATA--ECRG----TTHT------QA--ACAT--WA +-4 +6z2y_B ! ! ITLTRTPT----KT--HTETPAQA--EARC----PT--------KALTNT--VATA L ! +------------------3oym_A +-----5 ----VT--PT----------PAQAAGEARCQG------------------------ L ! ! +----------3jca_E ! +---------3 --------QTDT--TA------QG--ECSCKG------------------------ --6 ! +6vdk_A ! +---------2 ----------DT----------QG--ECSGHCQT---------------------- ! +6rwm_A ! --------------------PAQGAGEARGQC------------------------ +--------------------------5m0q_A |
home
updated Tue Dec 19 15:27:18 2023
