| structure name | ISCTH4 TRANSPOSASE, PRE-CLEAVED COMPLEX, PCC |
| reference | Kosek et al. 'Embo 40 e105666 2021 |
| experiment | X-ray (resolution=3.50, R-factor=0.222) |
| structural superfamily | Ribonuclease H-like; |
| sequence family | Transposase, Mutator family; MULE transposase domain; |
| multimeric complexes | 6xg8_AB 6xgw_AB |
| redundant complexes |  6xgw_A 6xgw_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | YYNTRRYRTRSAESTVP |
Estimated binding specificities ?
| readout + contact | 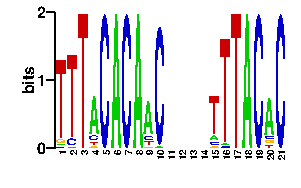 |
A | 0 5 0 0 96 87 73 24 24 24 24 0 11 0 0 0 0 10 96 79 87 C | 0 8 0 0 0 2 10 24 24 24 24 0 8 0 0 0 0 8 0 2 5 G | 96 7 96 0 0 5 8 24 24 24 24 93 13 0 96 0 96 11 0 10 2 T | 0 76 0 96 0 2 5 24 24 24 24 3 64 96 0 96 0 67 0 5 2scan! |
Dendrogram of similar interfaces ?
matrix formatYAYTNCTTRTRGYTRTTGRGSTATEAST------------------------TGVCPG L +-----------------------6xg8_A +--6 ----------------------------------------NGRGTGLGLCRG------ ! +-----------------------4ew0_A ! ------------RA--------------ITQTNAMG---------------------- L ! +5zjt_E +-7 +--------------1 ------------RT--------------IAQTNA------------------------ L ! ! ! +5lux_K ! ! +-4 ------------RT--------------VAQTNA------------------------ L ! ! ! ! +3lnq_A +-8 +----------5 +--------------2 ----------------------------IAQCNAMG---------------------- L ! ! ! +1b72_A ! ! ! ------------RA--------------IAQTNAMC---------------------- L ! ! +---------------1puf_A --9 ! ------------------------------------KT-------------------- L ! +----------------------------5ejk_E ! --------------------------------------LTNGRGTGLCLCRG------ ! +4ofa_A +---------------------------3 ----------------------------------------NGRGTGLGLCRG------ +4e9f_A |
home
updated Sat Jan 10 13:07:53 2026
