| structure name | CRYO-EM STRUCTURE OF THE HERMES TRANSPOSASE BOUND TO TWO LEFT-ENDS OF ITS DNA TRANSPOSON |
| experiment | NMR |
| structural superfamily | Ribonuclease H-like;Hermes dimerisation domain;beta-beta-alpha zinc fingers; |
| sequence family | Hermes transposase DNAbinding domain; hAT family dimerisation domain; BED zinc finger; |
| multimeric complexes | 8edg_CGK 8sjd_AC 8sjd_BD |
| redundant complexes |  8eb5_A
8sjd_A 8eb5_A
8sjd_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RQTSNRSW |
Estimated binding specificities ?
| contact | 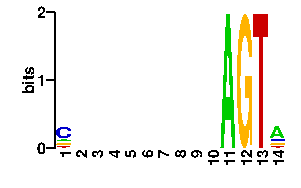 |
A | 13 24 24 24 24 24 24 24 24 24 96 0 0 54 C | 54 24 24 24 24 24 24 24 24 24 0 0 0 13 G | 13 24 24 24 24 24 24 24 24 24 0 96 0 16 T | 16 24 24 24 24 24 24 24 24 24 0 0 96 13scan! |
home
updated Sat Jan 10 23:09:39 2026
