| structure name | STRUCTURE OF MITOCHONDRIAL RNA POLYMERASE ELONGATION COMPLEX (DNA-DIRECTED RNA POLYMERASE, MITOCHONDRIAL) |
| reference | Schwinghammer et al. Nat.Struct.Mol.Biol. 20 1298 2013 |
| source | HOMO SAPIENS |
| experiment | X-ray (resolution=2.65, R-factor=0.181) |
| structural superfamily | DNA/RNA polymerases; |
| sequence family | DNAdependent RNA polymerase; |
| multimeric complexes | 6erp_ACF 6erp_BGJ 6erq_ACF 6erq_BGJ |
| redundant complexes |  6erq_B
6erp 6erq_B
6erp
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QSRISEQTYVYQWER |
Estimated binding specificities ?
| contact | 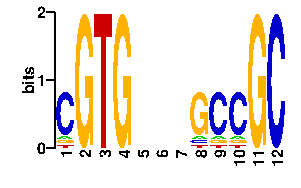 |
A | 96 0 0 96 12 10 24 24 96 0 85 C | 0 85 0 0 64 10 24 24 0 0 1 G | 0 11 96 0 10 66 24 24 0 96 0 T | 0 0 0 0 10 10 24 24 0 0 10scan! |
Dendrogram of similar interfaces ?
matrix formatQGSC--RG--------IC--------SC----EG----------QG--TGYGVG--YT--QG--WGEG--RG-------- L +-----------4boc_A +--5 ----YTHASAKT----------------------------------------------------------RG-------- L +-----------6 +-----------6ymv_A ! ! ------------------ATYTDGKG------------RCRCYC----TTYT----FART--IG------------HT-- L ! +--------------1msw_D ! ----------------------------KC----------RT--TC----YCMCSAHA----------------QG---- ! +-6fbc_A +-8 +-3 ----------------------------------------------FC--YG--------------------NGQC---- L ! ! ! +-1tau_A ! ! +----------4 ----------------------------KT----YG----RG----YG--YAIGSCYC----------RC----QA--HG ! ! ! ! +-4yfu_D ! ! ! +-2 ------------------------------RC--------RA----YG--YT--SCYC---------------------- L --9 +-----------7 ! +6vdd_A ! ! +-1 ------------------------------------VC------------------------------------------ ! ! +1kln_A ! ! ------------------------------------------RT------YA----------------------QA---- ! +--------------1x9m_A ! ------------IAVA----------------------RC--YC------------------------------------ +----------------------------6dta_A |
home
updated Sat Jan 10 06:54:07 2026
