| structure name | STRUCTURE OF THE CLR-CAMP-DNA COMPLEX (Putative cAMP-binding protein-catabolite gene activator) |
| reference | Essen et al. 'To ? ? ? |
| source | Sinorhizobium meliloti 1021 |
| experiment | X-ray (resolution=2.72, R-factor=0.272) |
| structural superfamily | cAMP-binding domain-like;"Winged helix" DNA-binding domain; |
| sequence family | Cyclic nucleotidebinding domain; Bacterial regulatory proteins, crp famil; |
| redundant complexes |  7pzb_A 7pzb_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | SRPKN |
Estimated binding specificities ?
| contact | 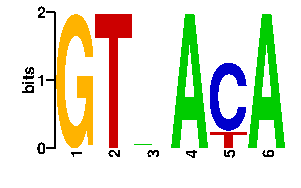 |
A | 0 0 39 96 0 96 C | 0 0 19 0 77 0 G | 96 0 19 0 0 0 T | 0 96 19 0 19 0scan! |
Dendrogram of similar interfaces ?
matrix formatKTQTSTRTETTA--RA------ L +--3iyd_H +--------------------------1 ----ITRGECSCSGKG------ L +-2 +--5i2d_R ! ! ------RTRGTT------KT-- L --3 +-----------------------------2o8k_A ! --------DCKT--RGSTRGYC L +-----------------------------4uno_A |
home
updated Sat Jan 10 17:09:24 2026
