| structure name | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS (PROTEIN SAP-1 ETS DOMAIN) |
| reference | Marmorstein et al. Mol.Cell 2 201 1998 |
| source | Homo sapiens |
| experiment | X-ray (resolution=1.93, R-factor=0.220) |
| structural superfamily | "Winged helix" DNA-binding domain; |
| sequence family | Etsdomain; |
| multimeric complexes | 1k6o_ABC |
| redundant complexes |  1bc7_C
1k6o_C 1bc7_C
1k6o_C
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | DKRARY |
Estimated binding specificities ?
| readout + contact | 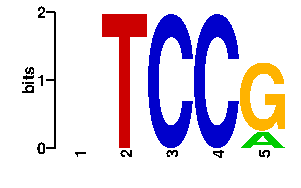 |
A | 19 0 0 0 20 C | 19 0 96 96 0 G | 19 0 0 0 76 T | 39 96 0 0 0scan! |
Dendrogram of similar interfaces ?
matrix format------QCKT--RGAT--RGNT-------- L +-----------------------------8smh_F ! ------DCKT--RG----RGYA--KA---- L +-3 +5jvt_A ! ! +--------------------1 ------DCKT--RGST--RGYA-------- L ! +-------2 +4bnc_A +-4 ! ----------RGNT--RGSTATRA------ L ! ! +---------------------2o6g_G +-5 ! ------NCTTNA------VTNT-------- L ! ! +-----------------------------1pp7_U --6 ! TT----KATTQTTG----LTRT--KAFTYT L ! +-----------------------------2p7c_B ! --KTHG------------------------ +-----------------------------5hdn_C |
home
updated Sat Jan 10 04:29:09 2026
