| structure name | FLP RECOMBINASE-HOLLIDAY JUNCTION COMPLEX I (FLP RECOMBINASE) |
| reference | Narendra et al. Mol.Cell 6 885 2000 |
| source | Saccharomyces cerevisiae |
| experiment | X-ray (resolution=2.65, R-factor=0.254) |
| structural superfamily | DNA breaking-rejoining enzymes;lambda integrase-like, N-terminal domain; |
| sequence family | Recombinase Flp protein; Recombinase Flp protein Nterminus; |
| redundant complexes |  1m6x_D
1p4e_A 1m6x_D
1p4e_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | ATSYTKRKNRSK |
Estimated binding specificities ?
| readout + contact | 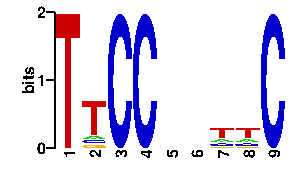 |
A | 0 42 33 24 24 0 0 96 96 C | 0 10 10 24 24 0 0 0 0 G | 96 21 21 24 24 96 96 0 0 T | 0 23 32 24 24 0 0 0 0scan! |
Dendrogram of similar interfaces ?
matrix format--AATA--YGTTKGRAFCKCKC------NT----RGST--KT L +--------------------1flo_A +----1 --KATANA------------KT--------TT--STRGYC-- +-2 +--------------------1z1b_B ! ! LTNTTT------------------TTQAATYG--YAKT---- L +-3 +-------------------------5jk0_B ! ! ------------------------------------RC---- L +-4 +---------------------------6en2_A ! ! ------------------------RT--ET------------ --5 +----------------------------6gys_E ! ----------------------KA------RTLT----RT-- L +----------------------------4e0p_A |
home
updated Tue Dec 19 05:38:13 2023
