| structure name | YEAST MATALPHA2/MCM1/DNA TERNARY TRANSCRIPTION COMPLEX CRYSTAL STRUCTURE (PROTEIN MAT ALPHA-2 TRANSCRIPTIONAL REPRESSOR) |
| reference | Richmond et al. |
| source | SACCHAROMYCES CEREVISIAE |
| experiment | X-ray (resolution=2.25, R-factor=0.240) |
| structural superfamily | Homeodomain-like; |
| sequence family | Homeobox domain; |
| multimeric complexes | 1mnm_ABCD |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RRNSNR |
Estimated binding specificities ?
| readout + contact | 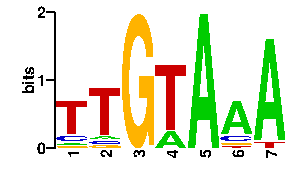 |
A | 6 1 0 30 96 63 89 6 C | 6 3 0 0 0 12 4 16 G | 16 3 96 0 0 6 0 9 T | 68 89 0 66 0 15 3 65scan! |
Dendrogram of similar interfaces ?
matrix format------NCICNARGRG L +-----------------------------4xrm_B +-3 RART--VTKGNA--KA L ! +-----------------------------2lkx_A --4 --RTQTVAQTNA---- L ! +8osb_E ! +---------------1 --RT--IAQCNAMG-- L +------------2 +1b72_A ! --RC--IAQCNAYC-- L +----------------3rkq_B |
home
updated Sun Aug 3 05:47:16 2025
