| structure name | CRYSTAL STRUCTURE OF THE MYCOBACTERIUM TUBERCULOSIS HYPOXIC RESPONSE REGULATOR DOSR C-TERMINAL DOMAIN-DNA COMPLEX (Dormancy Survival Regulator) |
| reference | Wisedchaisri et al. J.Mol.Biol. 354 630 2005 |
| source | Mycobacterium tuberculosis |
| experiment | X-ray (resolution=3.10, R-factor=0.272) |
| structural superfamily | C-terminal effector domain of the bipartite response regulators; |
| sequence family | Bacterial regulatory proteins, luxR family; |
| multimeric complexes | 1zlk_AB |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | KTKNYS |
Estimated binding specificities ?
readout + contact + contact | 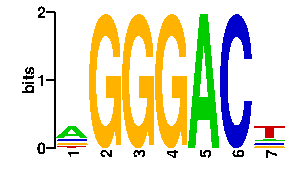 |
A | 54 0 0 0 96 0 13 C | 13 0 0 0 0 96 16 G | 13 96 96 96 0 0 13 T | 16 0 0 0 0 0 54scan! |
Dendrogram of similar interfaces ?
matrix format----------------YG---- +-----------------------------3eyi_B +-1 --SG--PTPGTT---------- L +-2 +-----------------------------7d98_Q ! ! TT----KATTQTTG--LTRTKA L --3 +-----------------------------2p7c_B ! --DCLT--AG--TTRG--RG-- L +-----------------------------5h3r_A |
home
updated Sat Jan 10 05:06:35 2026
