| structure name | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I LARGE FRAGMENT BOUND TO DUPLEX DNA WITH CYTOSINE-ADENINE MISMATCH AT (N-1) POSITION (DNA POLYMERASE I) |
| reference | Beese et al. |
| source | GEOBACILLUS |
| experiment | X-ray (resolution=1.53, R-factor=0.170) |
| structural superfamily | DNA/RNA polymerases;Ribonuclease H-like; |
| sequence family | DNA polymerase family A; |
| multimeric complexes | 8scg_AD 8sci_AD 8scj_AD 8sck_AD 8scl_AD 8scm_AD 8scn_AD 8sco_AD 8scp_AD 8scr_AD 8scs_AD 8sct_AD 8scu_AD |
| reference complex | 4yfu_D |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | KTYRNNYNQH |
Estimated binding specificities ?
readout + contact + contact | 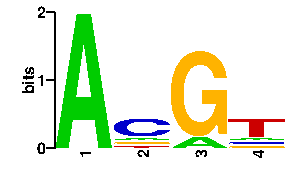 |
A | 96 12 12 12 C | 0 60 0 12 G | 0 12 84 12 T | 0 12 0 60scan! |
home
updated Thu Jan 8 23:11:55 2026
