| structure name | CRYSTAL STRUCTURE OF I-GZEII LAGLIDADG HOMING ENDONUCLEASE IN COMPLEX WITH DNA TARGET SITE (LAGLIDADG endonuclease) |
| reference | 'To ? ? ? X-RAY DIFFRACTION |
| source | Gibberella zeae |
| experiment | X-ray (resolution=2.80, R-factor=0.188) |
| structural superfamily | Homing endonucleases; |
| sequence family | LAGLIDADG endonuclease; |
| redundant complexes |  4z1x_B 4z1x_B
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | MHERRQKNEKRSSYYSSRSNHNAVR |
Estimated binding specificities ?
| readout + contact | 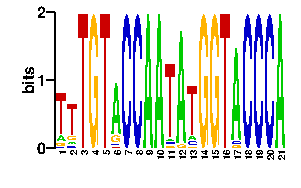 |
A | 0 0 0 0 0 96 0 0 67 0 81 0 0 0 0 4 96 0 96 64 67 C | 0 0 0 0 0 0 96 96 4 0 4 0 0 0 0 14 0 96 0 4 9 G | 0 96 96 96 0 0 0 0 9 0 4 0 0 96 96 4 0 0 0 14 9 T | 96 0 0 0 96 0 0 0 16 96 7 96 96 0 0 74 0 0 0 14 11scan! |
Dendrogram of similar interfaces ?
matrix formatSCMCRG----YTSTECTT--RG--TTRG--WTTTHGYCRGRGIT----QCKA----MCHTTGKT L +5thg_A +-----------------------1 SA--RARG----ECECKG--ST--ATRGRGWGCAFTQACT--SA--YTLA----FTWCEATT-- L ! +6uw0_B --3 SA--RGRGSTSTECQGTA--ST--AT--KG--CAFCQAQA----TAYAKG--KGFTWCEATA-- L ! +----------------4yhx_A +------2 SGIGQAKG--KG--KGTAYC--STVG--YA---------------------------------- L +----------------6fb0_B |
home
updated Sat Jan 10 06:57:30 2026
