| structure name | CRYSTAL STRUCTURE OF AN IDER-DNA COMPLEX REVEALS A CONFORMATIONAL CHANGE IN ACTIVATED IDER FOR BASE-SPECIFIC INTERACTIONS (Iron-dependent repressor ideR) |
| reference | Wisedchaisri et al. J.Mol.Biol. 342 1155 2004 |
| source | Mycobacterium tuberculosis |
| experiment | X-ray (resolution=2.75, R-factor=0.232) |
| structural superfamily | Iron-dependent repressor protein, dimerization domain;"Winged helix" DNA-binding domain;C-terminal domain of transcriptional repressors; |
| sequence family | Iron dependent repressor, metal bindin; Iron dependent repressor, Nterminal D; FeoA domain; MarR family; |
| multimeric complexes | 1u8r_ABCD 1u8r_GHIJ |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | SPTSQR |
Estimated binding specificities ?
| contact | 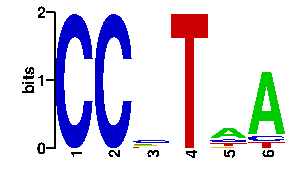 |
A | 0 0 19 0 52 78 C | 96 96 43 0 17 10 G | 0 0 17 0 10 0 T | 0 0 17 96 17 8scan! |
Dendrogram of similar interfaces ?
matrix formatTT----KATTQTTGLTRTKA--FTYT L +-----------------------------2p7c_B +-1 --------------YG---------- +-2 +-----------------------------3eyi_B ! ! --DCLT--AGTTRG--RG-------- L +-3 +-----------------------------5h3r_A ! ! ----RGSTRTSTHG------------ L --4 +-----------------------------3zpl_B ! --SG--PTPGTT--------HG---- L +-----------------------------7d98_Q |
home
updated Sun Aug 3 06:00:12 2025
