| structure name | CRYO EM STRUCTURE OF THE CONJUGATIVE RELAXASE TRAI OF THE F/R1 PLASMID SYSTEM (DNA HELICASE I) |
| source | ESCHERICHIA COLI |
| experiment | NMR |
| structural superfamily | Origin of replication-binding domain, RBD-like;P-loop containing nucleoside triphosphate hydrolases; |
| sequence family | TrwC relaxase; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | MIRHRQRRTPWTKSFKHREAITRDLHKM |
Estimated binding specificities ?
| contact | 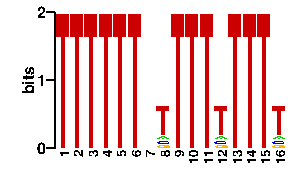 |
A | 0 0 0 0 0 0 24 9 0 0 0 9 0 0 0 9 C | 0 0 0 0 0 0 24 11 0 0 0 11 0 0 0 9 G | 0 0 0 0 0 0 24 9 0 0 0 9 0 0 0 11 T | 96 96 96 96 96 96 24 67 96 96 96 67 96 96 96 67scan! |
Dendrogram of similar interfaces ?
matrix format----------------------------VTETRTHTPTVT-- L +-----------------------------6sjf_D +-3 ----------------------------------DT----RT L ! +-----------------------------4n0o_E +-4 --------------KATT------------------------ L ! ! +--------2p7c_B ! +--------------------2 ------RGHTSTFT---------------------------- L +-5 +--------1vtn_C ! ! --SGAT------------------------------------ L ! ! +4rb3_D --6 +----------------------------1 ------------------KTPTPGYTRG-------------- L ! +4mtd_D ! TT---------------------------------------- +-----------------------------8pw0_A |
home
updated Sat Jan 10 08:28:24 2026
