| structure name | CRYSTAL STRUCTURE OF TRE/LOXLTR COMPLEX (Tre recombinase protein) |
| reference | Karpinski et al. 'Nucleic 45 9726 2017 |
| source | synthetic construct |
| experiment | X-ray (resolution=3.10, R-factor=0.211) |
| structural superfamily | DNA breaking-rejoining enzymes;lambda integrase-like, N-terminal domain; |
| sequence family | Phage integrase family; |
| multimeric complexes | 5u91_AB 5u91_EF |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | HTEMSKTQQCRMKRRRSYQR |
Estimated binding specificities ?
| readout + contact | 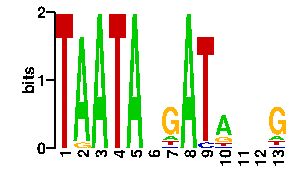 |
A | 0 96 96 0 96 24 11 96 5 56 24 24 11 C | 0 0 0 0 0 24 5 0 11 13 24 24 7 G | 0 0 0 0 0 24 67 0 1 16 24 24 73 T | 96 0 0 96 0 24 13 0 79 11 24 24 5scan! |
Dendrogram of similar interfaces ?
matrix formatHTTAECMAST--KTTTQTQT--CTRAMT--KA--RARTRCSA--YA--QG--------RA---- L +5u91_B +-------------------1 HTTTKGMTST--KATT--QT----QT----KA--RARTKTST--RG--EA--------RA---- +----2 +1xo0_B ! ! NATA----RTTT--NA--RASA--------------------ATVTDT--------RC--NTMT L +-4 +-------------------6en1_A ! ! ----------------------------------------TTQAATYGYAKT----------TT L +-5 +------------------------5jk0_B ! ! --------------------------------KT------------------TT---------- L --6 +--------------------------2b9s_A ! ----------------------------KC------------------RGST--KT-------- L ! +-----------------------1flo_A +---3 ----------------------------------------RT--ET------------------ +-----------------------6gys_E |
home
updated Wed Dec 18 02:23:30 2024
