| structure name | CRYSTAL STRUCTURE OF MOUSE MUTYH IN COMPLEX WITH DNA CONTAINING AP SITE ANALOGUE:8-OXOG (FORM II) (Adenine DNA glycosylase) |
| reference | Nakabeppu et al. 'Nucleic 49 7154 2021 |
| source | Mus musculus |
| experiment | X-ray (resolution=1.97, R-factor=0.179) |
| structural superfamily | DNA-glycosylase;Nudix; |
| sequence family | HhHGPD superfamily base excision DNA r; NUDIX domain; Helixhairpinhelix motif; Ironsulfur binding domain of endonucle; |
| redundant complexes |  7ef8_A 7ef8_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QQQTYSRRSH |
Estimated binding specificities ?
| contact | 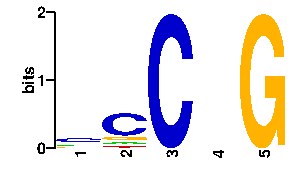 |
A | 16 10 0 24 0 C | 45 63 96 24 0 G | 16 13 0 24 96 T | 19 10 0 24 0scan! |
Dendrogram of similar interfaces ?
matrix format----------QG--QG----YC----------------RCSCHC L +---7ef9_A +-----------------2 ----------QG--RG----YC------------------SCHC ! +---6u7t_A +--6 --------------------YG----------ACCTRG------ ! ! +--------5kn8_A ! +------------4 ----------QTLAHGAT------KT------------------ +-7 +--------3s6i_A ! ! ----------QGCGTA---------------------------- ! ! +-------1orn_A ! ! +----3 ----------NG--RG------FGRC------------------ --8 +-----------5 +-------4ejy_A ! ! --------AGNG------RCFC----RG--RA------------ ! +------------3knt_A ! --------NGRGTG------LG--LCRG---------------- ! +7kz0_A +-------------------------1 LTVTQTLTNGRGTG------LC--LCRGYT-------------- +4ofa_A |
home
updated Sat Jan 10 15:02:29 2026
