| structure name | CRYO-EM STRUCTURE OF YEAST MITOCHONDRIAL RNA POLYMERASE TRANSCRIPTION INITIATION COMPLEX WITH TWO GTP MOLECULES POISED FOR DE NOVO INITIATION (IC2) |
| experiment | NMR |
| structural superfamily | S-adenosyl-L-methionine-dependent methyltransferases; |
| sequence family | Ribosomal RNA adenine dimethylase; |
| multimeric complexes | 6ymv_AB 6ymw_AB 8ap1_AB 8att_AB 8atv_AB 8atw_AB 8c5s_AB 8c5u_AB 8q63_AB |
| reference complex | 6ymv_B |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | FYWELMQYQ |
Estimated binding specificities ?
| contact | 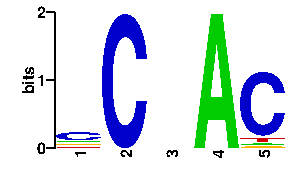 |
A | 15 0 24 96 4 C | 51 96 24 0 81 G | 15 0 24 0 4 T | 15 0 24 0 7scan! |
home
updated Fri Jan 9 20:40:01 2026
