| structure name | STRUCTURE OF NFAT1 BOUND AS A DIMER TO THE HIV-1 LTR KB ELEMENT (Nuclear factor of activated T-cells, cytoplasmic 2) |
| reference | von Koenig et al. Nat.Struct.Biol. 10 800 2003 |
| source | Homo sapiens |
| experiment | X-ray (resolution=2.60, R-factor=0.231) |
| structural superfamily | p53-like transcription factors;E set domains; |
| sequence family | Rel homology domain RHD; IPT/TIG domain; |
| multimeric complexes | 1a02_FJN 1p7h_LN 1pzu_BHIM 1s9k_CDE 2o93_LMO |
| redundant complexes | 1owr_P
1pzu_M
2as5_N
2o93_O
 8ow4_B
1a02
1s9k
3qrf 8ow4_B
1a02
1s9k
3qrf
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RYTERRQKR |
Estimated binding specificities ?
| contact | 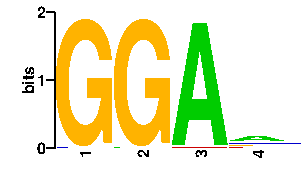 |
A | 0 1 94 47 C | 1 0 0 17 G | 95 95 0 16 T | 0 0 2 16scan! |
Dendrogram of similar interfaces ?
matrix formatRGRGYTECHG------ L +-----------------------------2o61_B ! ------------PTRG L --3 +1nfk_A ! +--------------1 --------------RG L +--------------2 +1svc_P ! RGRGYTECHGKG---- L +--------------1a3q_A |
home
updated Sat Jan 10 04:52:37 2026
