| structure name | K103A/K262A DOUBLE MUTANT OF I-SMAMI (I-SmaMI LAGLIDADG meganuclease) |
| reference | Stoddard et al. J.Mol.Biol. 428 206 2016 |
| source | Sordaria macrospora (strain ATCC MYA-333 / DSM 997 / K(L3346) / K-hell) / Sordaria macrospora (strain ATCC MYA-333 / DSM 997 / K(L3346) / K-hell) |
| experiment | X-ray (resolution=2.20, R-factor=0.183) |
| structural superfamily | Homing endonucleases; |
| sequence family | LAGLIDADG endonuclease; |
| redundant complexes | 4z1z_A
4z20_D
 5e5o_A 5e5o_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | SMRRKYLVSTKSTSREKIKSIQIQTEDRWYT |
Estimated binding specificities ?
| readout + contact | 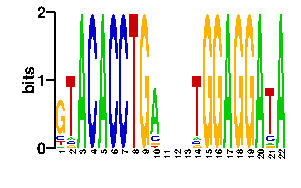 |
A | 5 5 96 0 96 0 0 0 0 74 24 24 24 5 0 0 96 0 0 96 7 96 C | 12 6 0 96 0 96 96 0 0 12 24 24 24 5 0 0 0 0 0 0 10 0 G | 69 5 0 0 0 0 0 0 96 5 24 24 24 5 96 96 0 96 96 0 5 0 T | 10 80 0 0 0 0 0 96 0 5 24 24 24 81 0 0 0 0 0 0 74 0scan! |
Dendrogram of similar interfaces ?
matrix formatSGMGRGRG--KTYALT--VT----TT----KT--SC----TA--RGEC----KGITKG--SGITQAICQATC--ECDTRG--WTYC--TC L +5e67_A +--------------------2 ST--SC--------HT--RT--TTMT----LT--NC----YA----YAWGSAFCRGRG------RGQGECSCYT------RTWCQTRTEC L ! +7rcf_A +------4 STITQA----ST--RG--RG--YCTCYC----RG------VCQT------STITQAKGRGSTNTQT--ATTC----YTSC--VCDTRGST L ! ! +--------------3mx9_A ! +------3 CA--QGVTRTDCYA----RT----FG----RG--RG--DTMCEA---------------------------------------------- L --5 ! +5a78_B ! +--------------1 STITQAKGRGSTNTQT--ST--ATTC----YT----SC--VCDTRGST------------------------------------------ L ! +3mxb_B ! --YTLC--------RGSGECRGVTTG--QG--VG------RTEARGSC------------------------------------------ L +----------------------------5o6i_A |
home
updated Sat Jan 10 08:00:35 2026
