- Figure B1. Flowchart of the processing of PDB entries in the 3D-footprint database.
- Figure B2. Distribution of distances (Å) in water-mediated hydrogen bonds (January 2008).
- Figure B3. Distribution of distances (Å) in thymine C7 hydrophobic contacts (January 2008).
- Figure B4. Comparison of contact and readout PWMs (April 2009).
- Figure B5. Specificity of curated and structure-based PWMs (April 2009, Excel file here).
- Figure B6. 3D-footprint database scheme.
- Table B7. matrix-quality benchmark of Escherichia coli 3D-footprints (2010).
- Benchmark results from the 2008 paper
'Prediction of TF target sites based on atomistic models of protein-DNA complexes':
- Figure 1. Scatter plot and regression analysis of the scoring capability of atomic interaction tables.
- Table 2. Scoring of PurR[2pua] cognate binding sites using readout PWMs.
- Figure 3. ROC plots of cognate site recovery in a set of random sequences for NarL[1je8],CRP[1cgp],PurR[2pua] and DnaA[1j1v].
- Table 3 and Figure 4. Comparison of contact and readout PWMs for 4 prokaryotic and 4 eukaryotic transcription factors.
- Figure 5. Stacked sequence logos for comparative models of transcription factors FNR and Giant.
- The 2011 paper 'Direct inference of protein-DNA interactions using compressed sensing methods' by M.AlQuraishi and H.H.McAdams provides an external benchmark of the DNAPROT algorithm with 63 non-redundant Helix-Turn-Helix proteins.
- The 2013 paper 'PiDNA: predicting protein–DNA interactions with structural models' by Chih-Kang Lin and Chien-Yu Chen compares the DNAPROT algorithm to PiDNA and 3DTF.
-
Figure B1.
Flowchart of the process applied to entries in the 3D-footprint database.
-
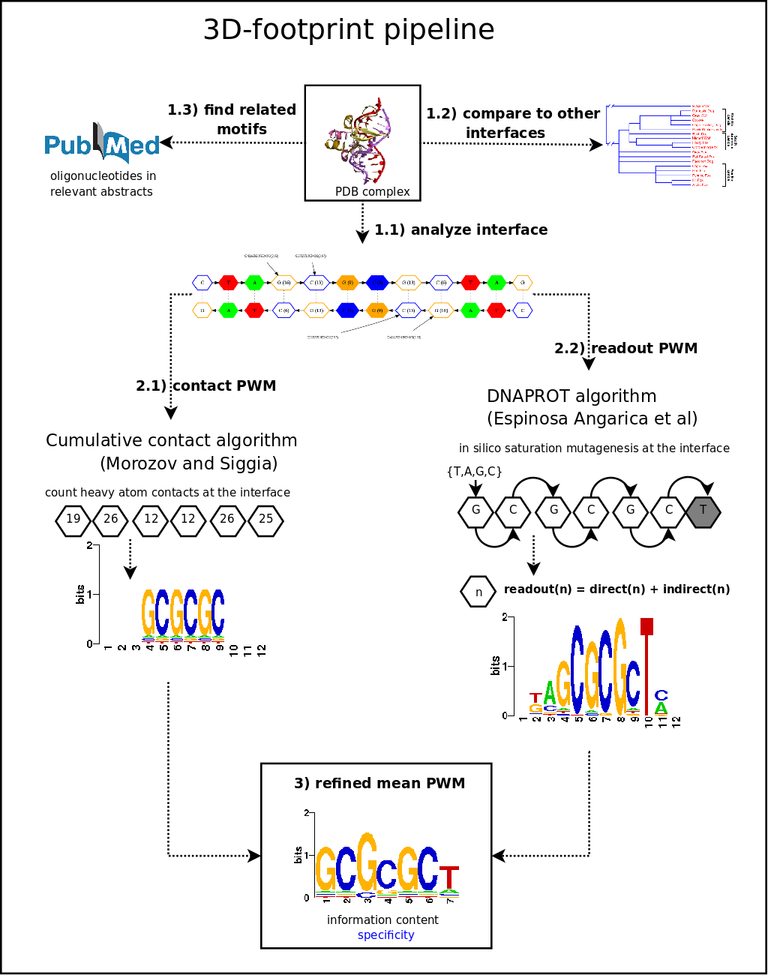
Figure B2.
Distribution of distances (Å) in water-mediated hydrogen bonds (January 2008).
-
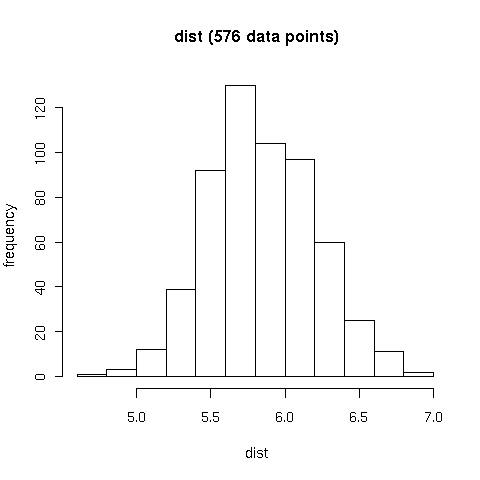
Figure B3.
Distribution of distances (Å) in thymine C7 hydrophobic contacts (January 2008).
-
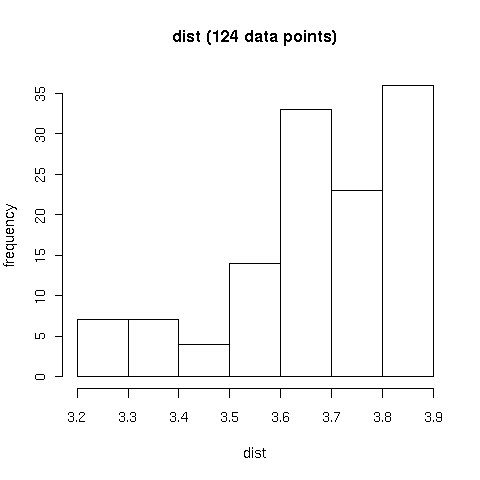
Figure B4.
A) Comparison of contact and readout positions weight matrices (PWMs) by means of
STAMP global alignments,
with similarity computed as the expectation value (Evalue).
B) Information content (IC) of readout PWMs calculated with the DNAPROT algorithm as
a function of the observed atomic contacts at the interface of protein-DNA complexes.
-
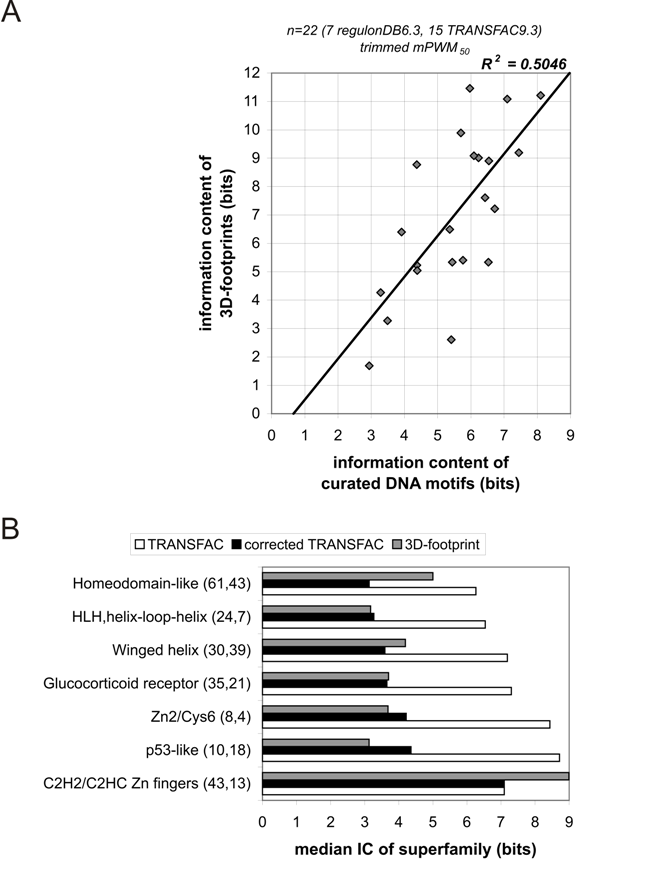
Figure B5.
A) Motif IC (specificity) of 22 transcription factors (7 in RegulonDB version 6.3 and 15 in TRANSFAC v9.3) which had protein-DNA complexes
available in the Protein Data Bank by 30 April 2009, compared to the IC associated to structure-based PWMs.
B) Median IC of several SCOP superfamilies as measured from 3D-footprints and from TRANSFAC PWMs, curated from the literature by expert curators.
The numbers in parenthesis indicate the number of PWMs in each superfamily in TRANSFAC and 3D-footprint, respectively.
In both panels TRANSFAC and regulonDB PWMs were trimmed/corrected to match the length of 3D-footprints if the difference in length was over 40%.
-
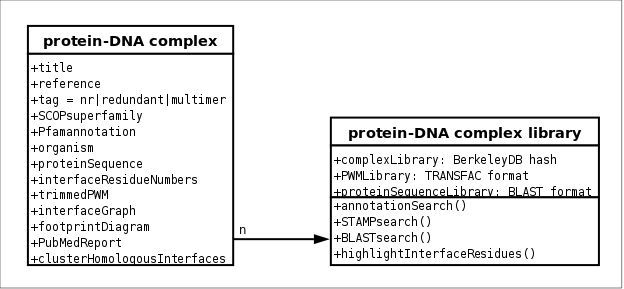
Figure B6.
Scheme of 3D-footprint database, that includes libraries of complexes, structure-based PWMs and protein sequences, and hence supports
three types of queries: annotation query (powered by BerkeleyDB and
CPAN::DB_File),
PWM query (powered by STAMP) and protein sequence query
(powered by BLAST).
-
Table B7.
matrix-quality
reports produced by my colleague A.Medina-Rivera on a subset of 3D-footprint PWMs derived for Escherichia coli
transcription factors (TFs). Each PWM is evaluated in the context of the complete E.coli K12 genome and compared to the set of DNA operator
sites for the corresponding TF annotated at RegulonDB (labelled as 'db_sites' in the charts).
This experiment considered PWMs time-stamped on 11/03/2010 and followed the protocol described in the paper
'Theoretical and empirical quality assessment of transcription factor-binding motifs'.
protein name | 3D-footprint entry | matrix-quality report Ada 1zgw_A Ada_1zgw_A CRP 1cgp_AB CRP_1cgp_AB DnaA 1j1v_A DnaA_1j1v_A FadR 1hw2_AB FadR_1hw2_AB LacI 1efa_AB LacI_1efa_AB MarA 1bl0_A MarA_1bl0_A NarL 1je8_AB NarL_1je8_AB PhoB 1gxp_AB PhoB_1gxp_AB PurR 2pua_A PurR_2pua_A PutA 2rbf_AB PutA_2rbf_AB Rob 1d5y_AB Rob_1d5y_AB TrpR 1rcs_AB TrpR_1rcs_AB
