| structure name | CRYSTAL STRUCTURE OF A CREB BZIP-CRE COMPLEX REVEALS THE BASIS FOR CREB FAIMLY SELECTIVE DIMERIZATION AND DNA BINDING (TRANSCRIPTION FACTOR CREB) |
| reference | Schumacher et al. J.Biol.Chem. 275 35242 2000 |
| source | Mus musculus |
| experiment | X-ray (resolution=3.00, R-factor=0.218) |
| structural superfamily | Leucine zipper domain; |
| sequence family | bZIP transcription factor; Basic region leucine zipper; |
| multimeric complexes | 1dh3_AC 5zk1_AC 5zko_ACGH |
| redundant complexes |  5zko_C
5zk1 5zko_C
5zk1
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | NAASR |
Estimated binding specificities ?
| contact | 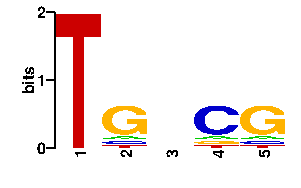 |
A | 0 11 24 9 9 C | 0 9 24 67 9 G | 0 67 24 9 69 T | 96 9 24 11 9scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: CREB | ||
| TGACGTCA | PubMed | 1dh3_C(1.78e-03), 1dh3_AC(2.83e-10), 5zko_ACGH(6.61e-12) |
| TGACG | PubMed | 1dh3_C(1.35e-04), 1dh3_AC(1.76e-06), 5zk1_AC(1.02e-03), ... (total=4) |
| CGTCA | PubMed | 1dh3_C(1.35e-04), 1dh3_AC(1.76e-06), 5zk1_AC(1.02e-03), ... (total=4) |
| TGACATCA | PubMed | 1dh3_AC(1.26e-05), 5zko_ACGH(5.71e-07) |
| TGACGT | PubMed | 1dh3_C(3.57e-04), 1dh3_AC(1.88e-05), 5zk1_AC(3.41e-03), ... (total=4) |
| CGGAAGTGACGTCAC | PubMed | 1dh3_C(1.25e-02), 1dh3_AC(2.06e-07), 5zko_ACGH(3.17e-06) |
| ATGACGTCAT | PubMed | 1dh3_C(4.09e-03), 1dh3_AC(8.38e-09), 5zko_ACGH(5.16e-09) |
| GTGACGTAC | PubMed | 1dh3_C(2.94e-03), 1dh3_AC(1.58e-03), 5zko_ACGH(2.67e-04) |
| AGATGATGTAA | PubMed | 5zko_ACGH(2.32e-03) |
| TGACCTCA | PubMed | 1dh3_AC(1.26e-05), 5zko_ACGH(5.71e-07) |
| TGAGGTCA | PubMed | 1dh3_AC(1.26e-05), 5zko_ACGH(5.71e-07) |
| GTCAT | PubMed | 1dh3_AC(1.77e-03), 5zko_ACGH(7.13e-03) |
| TAACGTTA | PubMed | 1dh3_AC(1.31e-02), 5zko_ACGH(1.23e-03) |
| CGGAAGTGACGTCA | PubMed | 1dh3_C(1.01e-02), 1dh3_AC(1.22e-07), 5zko_ACGH(1.40e-06) |
| TGGCG | PubMed | 1dh3_C(1.35e-04), 1dh3_AC(1.87e-03) |
| site | source | matches (E-value) |
Dendrogram of similar interfaces ?
matrix format------RA----NA--AAVT--STRG-- L +1h89_A +-1 ------------RG----NC--YGATRC L +-----------------3 +4eot_A ! ! ----RG------NC--YGAT--CTRG-- L ! +2wt7_B ! ------------NT--ATAT--STRG-- L ! +1dh3_C +-------8 +---4 RA----NA----AAVT--STRG------ L ! ! ! ! +1nwq_C ! ! +-6 +-2 --AAVT----STRG-------------- L ! ! ! ! +1hjb_A ! ! ! ! --QC----FTRC---------------- L --9 +-----------7 +----7aw7_D ! ! ------NC----AT----STRA------ ! ! +-1a02_J ! +----5 ------------NCTT--AT----RG-- L ! +-2dgc_A ! ----NCRT--ATAT--STRG-------- +--------------------------1t2k_D |
home
updated Sat Jan 10 04:33:21 2026
