| structure name | CRYSTAL STRUCTURE OF TFAM (MITOCHONDRIAL TRANSCRIPTION FACTOR A) IN COMPLEX WITH LSP (Transcription factor A, mitochondrial) | ||||
| reference | Garcia-Diaz et al. 'Nucleic 50 322 2022 | ||||
| source | Homo sapiens | ||||
| experiment | X-ray (resolution=2.70, R-factor=0.210) | ||||
| structural superfamily | HMG-box; | ||||
| sequence family | HMG high mobility group box; | ||||
| multimeric complexes | 6erp_ACF 6erp_BGJ 6erq_ACF 6erq_BGJ 7lbw_AB | ||||
| redundant complexes |  7lbw_B
6erp
6erq 7lbw_B
6erp
6erq
| ||||
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| contact | 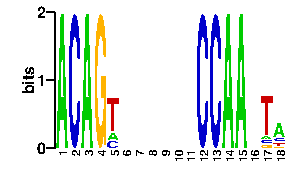 |
A | 0 0 0 0 0 13 24 24 24 24 24 24 13 0 0 0 0 C | 0 0 0 0 0 16 24 24 24 24 24 24 54 0 0 0 0 G | 96 0 0 96 96 54 24 24 24 24 24 24 13 0 96 0 0 T | 0 96 96 0 0 13 24 24 24 24 24 24 16 96 0 96 96scan! |
Dendrogram of similar interfaces ?
matrix format--STYGLCSGLAKTTGTCIGMGKGRTYANTVTFGPGQCLC L +---------------7lbx_A +------1 RCNTFTMA--RAHGNAACST-------------------- L +--2 +---------------4euw_A ! ! RTNTFTMA--RT--NG--SA-------------------- L +-3 +----------------------4y60_C ! ! KCNT--MAMA--EGSAACNT-------------------- L +-4 +-------------------------2lef_A ! ! ------FTCA----IG--AT-------------------- +-5 +--------------------------6oem_H ! ! RGNTFTMA--RT--NGRTSA-------------------- L --6 +---------------------------7m5w_A ! ----YA--------FT--SG-------------------- +---------------------------5ze0_N |
home
updated Sun Aug 3 16:56:27 2025
