| structure name | CRYSTAL STRUCTURE OF S. AUREUS GYRASE IN COMPLEX WITH 4-[[1-[(1- CHLORO-6,7-DIHYDRO-5H-CYCLOPENTA[C]PYRIDIN-6-YL)METHYL]AZETIDIN-3- YL]METHYLAMINO]-6-FLUOROCHROMEN-2-ONE |
| reference | 'Acs 2023 |
| experiment | X-ray (resolution=2.16, R-factor=0.191) |
| structural superfamily | Type II DNA topoisomerase; |
| sequence family | DNA gyrase/topoisomerase IV, subunit A; DNA gyrase B subunit, carboxyl terminus; Toprim domain; |
| multimeric complexes | 2xcr_BD 2xcr_SU 2xcs_BD 4bul_AC 4plb_BD 5bs3_BD 5cdo_ABCD 5cdo_RSTU 5cdq_ABCD 5cdq_RSTU 5cdr_AB 5cdr_CD 5iwi_AB 5iwi_CD 5iwm_ABCD 5npk_bd 6fm4_BD 6fqm_ABCD 6fqm_abcd 6fqv_ABCD 6fqv_RSTU 6qtk_ABCD 6qtp_ABCD 6qx1_ABCD 7fvs_AB 7fvt_AB 7mvs_AB |
| reference complex | 5iwi_C |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | KYRSSRI |
Estimated binding specificities ?
| contact | 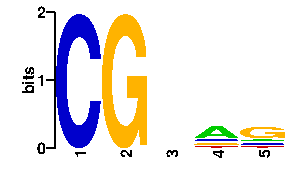 |
A | 16 13 24 0 0 C | 54 16 24 96 0 G | 13 13 24 0 96 T | 13 54 24 0 0scan! |
home
updated Fri Jan 9 12:44:22 2026
