| structure name | STRUCTURE OF NO-DNA COMPLEX (Nucleoid occlusion factor SlmA) |
| reference | Proc.Natl.Acad.Sci.USA 110 10586 2013 |
| source | Klebsiella pneumoniae |
| experiment | X-ray (resolution=2.05, R-factor=0.218) |
| structural superfamily | Homeodomain-like;Tetracyclin repressor-like, C-terminal domain; |
| sequence family | Bacterial regulatory proteins, tetR family; |
| multimeric complexes | 4gck_ABCD 4gcl_ABCD 4gcl_EGH 5hbu_ABCD 5hbu_EGH 5k58_ABEF |
| redundant complexes |  4gcl_C
5hbu_G
5k58_B 4gcl_C
5hbu_G
5k58_B
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | TSEAAYR |
Estimated binding specificities ?
readout + contact + contact | 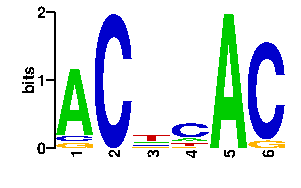 |
A | 0 0 17 43 0 10 C | 0 0 17 17 0 0 G | 96 0 54 17 96 8 T | 0 96 8 19 0 78scan! |
Dendrogram of similar interfaces ?
matrix formatTA--STECAGATYTRG L +4gck_A +-1 TA----ECAC--YCRA +-2 +5haw_A ! ! VT--AGDT--TTYTLA L +-------3 +5gpc_B ! ! ----TTRG----YTWA L +------4 +-7jnp_A ! ! LC--RTPTPTSTYT-- L ! +---------6nsn_B --7 LTRG--PTAC--YT-- L ! +---------3lsr_A ! +--5 ----TGKG----YCFA L +---6 +---------4pxi_B ! MTRGTGPT--ATYTWT L +------------5yej_B |
home
updated Sat Jan 10 07:04:48 2026
