| structure name | CRYO-EM STRUCTURE OF YEAST MITOCHONDRIAL RNA POLYMERASE PARTIALLY- MELTED TRANSCRIPTION INITIATION COMPLEX (PMIC) |
| experiment | NMR |
| structural superfamily | DNA/RNA polymerases; |
| sequence family | DNAdependent RNA polymerase; |
| multimeric complexes | 6ymv_AB 6ymw_AB 8ap1_AB 8att_AB 8atv_AB 8atw_AB 8c5s_AB 8c5u_AB 8q63_AB |
| redundant complexes | 8atw_A
 8q63_A
6ymw
8ap1
8att
8atv
8c5s
8c5u 8q63_A
6ymw
8ap1
8att
8atv
8c5s
8c5u
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | YHSKQFR |
Estimated binding specificities ?
| contact | 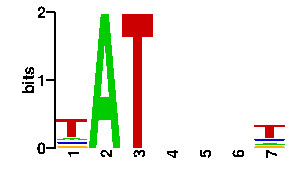 |
A | 13 27 24 24 96 0 60 C | 13 22 24 24 0 0 12 G | 56 25 24 24 0 0 12 T | 14 22 24 24 0 96 12scan! |
Dendrogram of similar interfaces ?
matrix format--------------------------YTHASAKT--------------------------------------QAFTRG------ L +-----------6ymv_A +----------1 --------QG--------------SC--RG----IC----------SC----EG------------QGTGYG----RG------ L +-----2 +-----------4boc_A ! ! ------------------------------------RGATYTDGKG----------RCRCYC------------------HT-- L +-3 +----------------------1msw_D ! ! ------------------------------------------------KT----YG--RG--NG--------------QA--HG +-4 +----------------------------4yfu_D ! ! KTATFCRC--DCSCWCRAITNTNG------------------------------------------------------------ L +-5 +-----------------------------4lup_A ! ! ------------------------------------------------KC----------RT---------------------- --6 +-----------------------------6fbc_A ! --------------------------------------------------RC------RA--NGNG------------------ L +-----------------------------6vdd_A |
home
updated Sat Jan 10 13:12:46 2026
