| structure name | CRYSTAL STRUCTURE OF THE MUTANT D75N I-CREI IN COMPLEX WITH ITS WILD- TYPE TARGET IN PRESENCE OF CA AT THE ACTIVE SITE (THE FOUR CENTRAL BASES, 2NN REGION, ARE COMPOSED BY GTAC FROM 5' TO 3') (DNA ENDONUCLEASE I-CREI) |
| reference | Marenchino et al. 'Nucleic 40 6936 2012 |
| source | CHLAMYDOMONAS REINHARDTII |
| experiment | X-ray (resolution=2.80, R-factor=0.181) |
| structural superfamily | Homing endonucleases; |
| sequence family | LAGLIDADG endonuclease; |
| multimeric complexes | 1bp7_AB 1bp7_CD 1g9y_AB 1t9i_AB 1t9j_AB 1u0c_AB 1u0d_AB 2i3p_AB 2i3q_AB 4aad_AB 4aae_AB 4aaf_AB 4aag_AB 4aqu_AB 6fb8_AB |
| reference complex | 1t9i_B |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | SIQKNSYQQTRRVKT |
Estimated binding specificities ?
| readout + contact | 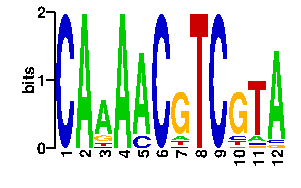 |
A | 0 96 69 91 82 1 6 0 0 4 8 87 C | 96 0 6 0 13 91 8 0 96 10 6 0 G | 0 0 8 4 1 0 74 0 0 69 4 5 T | 0 0 13 1 0 4 8 96 0 13 78 4scan! |
home
updated Thu Jan 8 23:49:38 2026
